This post presents basic R code snippets to read files with given file extensions such as csv or txt. This is simple but very useful when it comes to the case where there are too many files to read manually.
If we have too many (i.e. 1000 files) csv files or its variants, it is impossible to read these files one by one manually.
For example, let’s assume that there are 6 files (USD.CSV, EUR.csv, CNY.TXT, AUD.txt, CNY2.ttxt, USD.CCSV) in a target directory. The first four files (csv, CSV, txt, TXT) are the files that we want to read. The contents of these files are straightforward because the output will show these contents in the later.
In this case, we can use list.files() R function to get names of these files with some certain file extensions.
list.files() function
list.files() is a built-in R function which returns a list of names of files with a given pattern.
list.files(path, pattern="\\.(csv|txt)$",
ignore.case = TRUE, full.names = FALSE)
In the above R command, “\\.(csv|txt)$” pattern specifies that 1) it is applied at the end of file name($), 2) multiple file extensions such as csv or txt file ((csv|txt)) are allowed but not for similar extensions such as ccsv or ttxt(\\.). csv and CSV or txt and TXT are allowed because case sensitivity is ignored (ignore.case = TRUE).
R code
The following R code is easy and self-contained: 1) reads each csv (CSV) or txt (TXT) files and make each data.frame separately and 2) reads and collects them into one data.frame.
#========================================================#
# Quantitative ALM, Financial Econometrics & Derivatives
# ML/DL using R, Python, Tensorflow by Sang-Heon Lee
#
# https://shleeai.blogspot.com
#--------------------------------------------------------#
# Basic R : read all csv files when these are so many
#========================================================#
graphics.off() # clear all graphs
rm(list = ls()) # remove all files from your workspace
# working directory
setwd("D:/SHLEE/blog/R/many_csv")
# directory where csv files are located
path<- file.path(getwd())
#-------------------------------------------------------
# make a list of all file names with csv or txt ext.
#-------------------------------------------------------
# $ : end of file name
# (csv|txt) : multiple file extentions
# \\. : avoid unwanted cases such as .ccsv
#-------------------------------------------------------
v.filename <- list.files(
path, pattern="\\.(csv|txt)$",
ignore.case = TRUE,
full.names = FALSE)
#-------------------------------------------------------
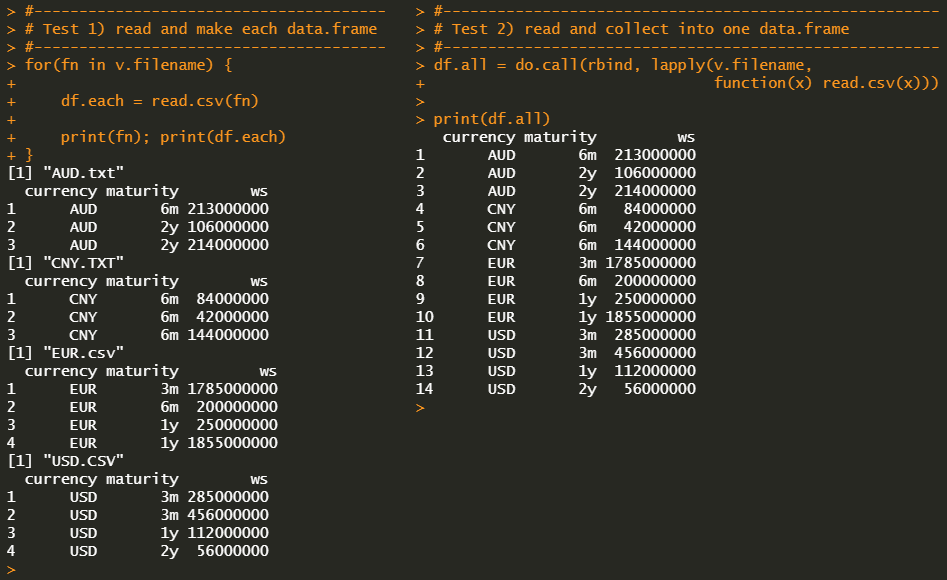
# Test 1) read and make each data.frame
#-------------------------------------------------------
for(fn in v.filename) {
df.each <- read.csv(fn)
# to do with df.each
# print
print(fn); print(df.each)
# use of assign()
#
# save each file as each data.frame
# which has each file name
assign(fn, read.csv(fn))
}
#-------------------------------------------------------
# Test 2) read and collect into one data.frame
#-------------------------------------------------------
df.all <- do.call(
rbind, lapply(v.filename,
function(x) read.csv(x)))
print(df.all)
# use of vroom package
library(vroom)
df.all.vroom <- vroom(v.filename)
print(df.all.vroom)
Output
We can find that only 4 files with correct file extensions are read while 2 unwanted files (.CCSV and .ttxt) are ignored.
Additional Output – assign() function
Josep Pueyo-Ros kindly advised me to use assign() function to save as a different data.frame with the name of the file. I think it is very useful so I add this function to R code and get the following 4 data.frames which can be seen at Environment/Data explorer in R studio.
- USD.CSV
- EUR.csv
- CNY.TXT
- AUD.txt
Additional Output – vroom package
HP (kind blog visitor) suggests the use of vroom which is a recently developed R package designed specifically for speed. I think this has lots of merits such as fast speed and suitability for complicated task. Result from vroom() function is as follows.
> df.all.vroom <- vroom(v.filename) Rows: 14 Columns: 3 -- Column specification ------------------- Delimiter: "," chr (2): currency, maturity dbl (1): ws i<U+00A0>Use spec() ( ) to retrieve the full column specification for this data. i<U+00A0>Specify the column types or set `show_col_types = FALSE` to quiet this message. > print(df.all.vroom) # A tibble: 14 x 3 currency maturity ws <chr> <chr> <dbl> 1 AUD 6m 213000000 2 AUD 2y 106000000 3 AUD 2y 214000000 4 CNY 6m 84000000 5 CNY 6m 42000000 6 CNY 6m 144000000 7 EUR 3m 1785000000 8 EUR 6m 200000000 9 EUR 1y 250000000 10 EUR 1y 1855000000 11 USD 3m 285000000 12 USD 3m 456000000 13 USD 1y 112000000 14 USD 2y 56000000 >
This R code is efficient and useful especially when there are too many files to read.
Originally posted on SHLee AI Financial Model blog.
Disclosure: Interactive Brokers
Information posted on IBKR Campus that is provided by third-parties does NOT constitute a recommendation that you should contract for the services of that third party. Third-party participants who contribute to IBKR Campus are independent of Interactive Brokers and Interactive Brokers does not make any representations or warranties concerning the services offered, their past or future performance, or the accuracy of the information provided by the third party. Past performance is no guarantee of future results.
This material is from SHLee AI Financial Model and is being posted with its permission. The views expressed in this material are solely those of the author and/or SHLee AI Financial Model and Interactive Brokers is not endorsing or recommending any investment or trading discussed in the material. This material is not and should not be construed as an offer to buy or sell any security. It should not be construed as research or investment advice or a recommendation to buy, sell or hold any security or commodity. This material does not and is not intended to take into account the particular financial conditions, investment objectives or requirements of individual customers. Before acting on this material, you should consider whether it is suitable for your particular circumstances and, as necessary, seek professional advice.














Join The Conversation
If you have a general question, it may already be covered in our FAQs. If you have an account-specific question or concern, please reach out to Client Services.